Here, we'll describe the bcgRNA library construction step-by-step. Other resources have described in detail library construction for 10x Genomics 3' scRNA-seq chemistry, and CITE-seq and Cell Hashing.
RfxCas13d will process CRISPR array into individual mature crRNAs ending with a single detacble barcoded gRNA:
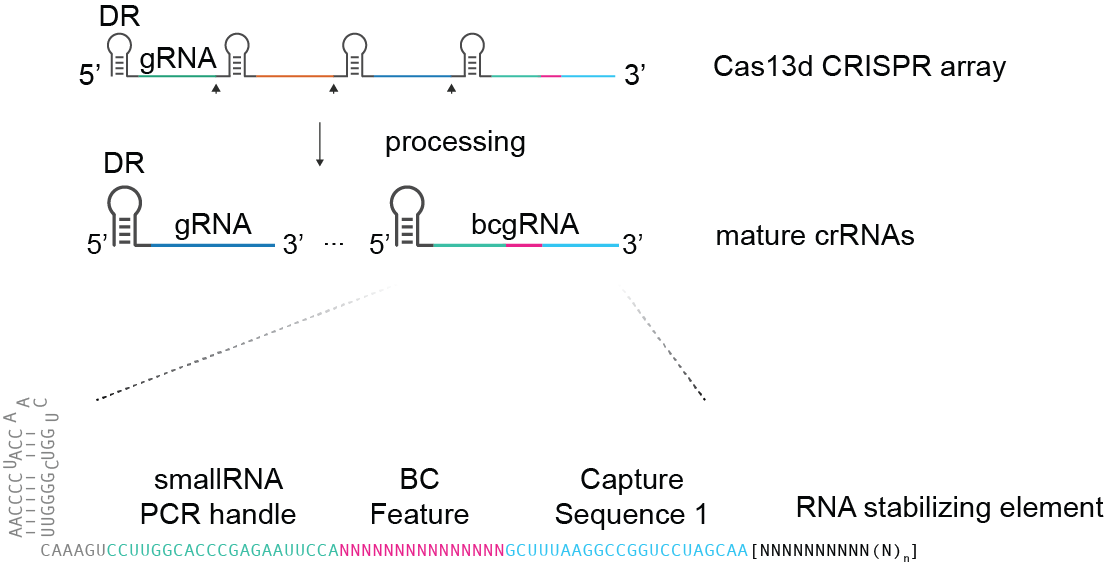
bcgRNA sequence:
10x V3 3' kit bead oligos (there are three types of sequences):
TruSeq Partial Read1 dT (for mRNA): |--5'- CTACACGACGCTCTTCCGATCT [16-bp cell barcode] [12-bp UMI] (T)30 -3' Nextera Partial Read1 Capture Sequence 1 (for surface protein & CRISPR screening): |--5'-GTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC -3' Nextera Partial Read1 Capture Sequence 2 (for CRISPR screening): |--5'-GTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] CCTTAGCCGCTAATAGGTGAGC -3'Feature cDNA Primers 1 (PN-2000096, for CRISPR screening) contain two primers for sgRNA/bcgRNA amplification.
To improve bcgRNA amplification, we add an additive reverse primer (not provided in PN-2000096), similar to CITE-seq:
Primers to amplify cDNA from bcgRNA: Forward primer: 5'-GCAGCGTCAGATGTGTATAAGAGACAG -3' Reverse primer: 5'-AAGCAGTGGTATCAACGCAGAG -3' Additive primer: 5'-CCTTGGCACCCGAGAATT*C*C -3'bcgRNA library amplification primers [We'll use the forward primer from Feature SI Primers 1 (PN-2000099)]:
Primers to index bcgRNA library: Forward primer: 5'-AATGATACGGCGACCACCGAGATCTACAC TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG -3' Reverse primer: 5'-CAAGCAGAAGACGGCATACGAGAT [8bp sample index]GTGACTGGAGTT CCTTGGCACCCGAGAATTCCA -3'Template Switching Oligo (TSO): 5'-
AAGCAGTGGTATCAACGCAGAGTACAT rGrGrG -3'Additive primer: 5'-
CCTTGGCACCCGAGAATT*C*C -3'Feature SI Primers 2 (PN-2000099): 5'-
AATGATACGGCGACCACCGAGATCTACAC TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG -3'RPI i7 Sample Index primer: 5'-
CAAGCAGAAGACGGCATACGAGAT [i7]GTGACTGGAGTT CCTTGGCACCCGAGAATTCCA -3'Illumina Nextera Read 1 primer: 5'-
TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG -3'Illumina smallRNA Read 2 primer: 5'-
GTGACTGGAGTT CCTTGGCACCCGAGAATTCCA -3'Illumina P5 adapter: 5'-
AATGATACGGCGACCACCGAGATCTACAC -3'Illumina P7 adapter: 5'-
CAAGCAGAAGACGGCATACGAGAT -3'Illumina P5 primer: 5'-
AATGATACGGCGACCACCGA -3'Illumina P7 primer: 5'-
CAAGCAGAAGACGGCATACGAGA -3'
bcgRNA:
If using Capture Sequence 1:
|--5'- GTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC -------->AACGAUCCUGGCCGGAAUUUCG NNNNNNNNNNNNNNN ACCUUAAGAGCCCACGGUUCC CAAAGUUUGGGGCUGGUCAACCAUCCCCAA -5' 3'- [n(N)NNNNNNNNNN]
|--5'- GTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG GTTTCAAACCCCGACCAGTTGGTAGGGGTT CCC -3'AACGAUCCUGGCCGGAAUUUCG NNNNNNNNNNNNNNN ACCUUAAGAGCCCACGGUUCC CAAAGUUUGGGGCUGGUCAACCAUCCCCAA -5' 3'- [n(N)NNNNNNNNNN]
Template switching may be inefficient due to terminal stem loop in template RNA.
|--5'- GTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG GTTTCAAACCCCGACCAGTTGGTAGGGGTT CCC-------->AACGAUCCUGGCCGGAAUUUCG NNNNNNNNNNNNNNN ACCUUAAGAGCCCACGGUUCC CAAAGUUUGGGGCUGGUCAACCAUCCCCAA GGGTACATGAGACGCAACTATGGTGACGAA -5' 3'- [n(N)NNNNNNNNNN]
(i) using PN-2000096 F+R primers
5'- GCAGCGTCAGATGTGTATAAGAGACAG ---------------> |--5'-GTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG GTTTCAAACCCCGACCAGTTGGTAGGGGTT CCCATGTACTCTGCGTTGATACCACTGCTT -3' <---------------TACATGAGACGCAACTATGGTGACGAA -5'
(ii) using PN-2000096 F and additive R primers (independent of template switching)
5'- GCAGCGTCAGATGTGTATAAGAGACAG ---------------> |--5'-GTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG GTTTCAAACCCCGACCAGTTGGTAGGGGTT CCCATGTACTCTGCGTTGATACCACTGCTT -3' <---------------CCTTAAGAGCCCACGGTTCC -5'
(i) cDNA amplification TSO product (173bp)
5'- GCAGCGTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG GTTTCAAACCCCGACCAGTTGGTAGGGGTT CCCATGTACTCTGCGTTGATACCACTGCTT -3' 3'-CGTCGCAGTCTACACATATTCTCTGTC [16-bp cell barcode] [12-bp UMI] AACGATCCTGGCCGGAATTTCG NNNNNNNNNNNNNNN ACCTTAAGAGCCCACGGTTCC CAAAGTTTGGGGCTGGTCAACCATCCCCAA GGGTACATGAGACGCAACTATGGTGACGAA -5'
(ii) cDNA amplification additive primer product (113bp) (Main product)
5'- GCAGCGTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG -3' 3'-CGTCGCAGTCTACACATATTCTCTGTC [16-bp cell barcode] [12-bp UMI] AACGATCCTGGCCGGAATTTCG NNNNNNNNNNNNNNN ACCTTAAGAGCCCACGGTTCC -5'
(i)
5'- AATGATACGGCGACCACCGAGATCTACAC TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG ---------------> 5'-GCAGCGTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG GTTTCAAACCCCGACCAGTTGGTAGGGGTT CCCATGTACTCTGCGTTGATACCACTGCTT -3' 3'-CGTCGCAGTCTACACATATTCTCTGTC [16-bp cell barcode] [12-bp UMI] AACGATCCTGGCCGGAATTTCG NNNNNNNNNNNNNNN ACCTTAAGAGCCCACGGTTCC CAAAGTTTGGGGCTGGTCAACCATCCCCAA GGGTACATGAGACGCAACTATGGTGACGAA -5' <---------------ACCTTAAGAGCCCACGGTTCC TTGAGGTCAGTG [i7]TAGAGCATACGGCAGAAGACGAAC -5’
(ii)
5'- AATGATACGGCGACCACCGAGATCTACAC TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG ---------------> 5'-GCAGCGTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG -3' 3'-CGTCGCAGTCTACACATATTCTCTGTC [16-bp cell barcode] [12-bp UMI] AACGATCCTGGCCGGAATTTCG NNNNNNNNNNNNNNN ACCTTAAGAGCCCACGGTTCC -5' <---------------ACCTTAAGAGCCCACGGTTCC TTGAGGTCAGTG [i7]TAGAGCATACGGCAGAAGACGAAC -5’
5'- AATGATACGGCGACCACCGA ---------------> 5'-AATGATACGGCGACCACCGAGATCT ACACTCGTCGGCAGCGTCAGATGTGTATAAGAGACAG [16-bp cell barcode] [12-bp UMI] TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG AACTCCAGTCAC [i7]ATCTCGTATGCCGTCTTCTGCTTG -3' 3'-TTACTATGCCGCTGGTGGCTCTAGA TGTGAGCAGCCGTCGCAGTCTACACATATTCTCTGTC [16-bp cell barcode] [12-bp UMI] AACGATCCTGGCCGGAATTTCG NNNNNNNNNNNNNNN ACCTTAAGAGCCCACGGTTCC TTGAGGTCAGTG [i7]TAGAGCATACGGCAGAAGACGAAC -5’ <---------------AGAGCATACGGCAGAAGACGAAC -5’
5'- AATGATACGGCGACCACCGAGATCT ACACTCGTCGGCAGCGTCAGATGTGTATAAGAGACAG NNNNNNNNNNNNNNNN NNNNNNNNNNNN TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG AACTCCAGTCAC NNNNNNNNATCTCGTATGCCGTCTTCTGCTTG -3' 3'-TTACTATGCCGCTGGTGGCTCTAGA TGTGAGCAGCCGTCGCAGTCTACACATATTCTCTGTC NNNNNNNNNNNNNNNN NNNNNNNNNNNN AACGATCCTGGCCGGAATTTCG NNNNNNNNNNNNNNN ACCTTAAGAGCCCACGGTTCC TTGAGGTCAGTG NNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5’
Illumina P5 Illumina Nextera Read1 Cell barcode UMI 10x CS1 Feature Barcode Illumina smRNA Read2 i7Illumina P7
(All primers are part of the Illumina sequencing kit.)
TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG ----------------------------> 3'-TTACTATGCCGCTGGTGGCTCTAGA TGTGAGCAGCCGTCGCAGTCTACACATATTCTCTGTC NNNNNNNNNNNNNNNN NNNNNNNNNNNN AACGATCCTGGCCGGAATTTCG NNNNNNNNNNNNNNN ACCTTAAGAGCCCACGGTTCC TTGAGGTCAGTG NNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5’
TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC -------> 3'-TTACTATGCCGCTGGTGGCTCTAGA TGTGAGCAGCCGTCGCAGTCTACACATATTCTCTGTC NNNNNNNNNNNNNNNN NNNNNNNNNNNN AACGATCCTGGCCGGAATTTCG NNNNNNNNNNNNNNN ACCTTAAGAGCCCACGGTTCC TTGAGGTCAGTG NNNNNNNNTAGAGCATACGGCAGAAGACGAAC -5’
5'- AATGATACGGCGACCACCGAGATCT ACACTCGTCGGCAGCGTCAGATGTGTATAAGAGACAG NNNNNNNNNNNNNNNN NNNNNNNNNNNN TTGCTAGGACCGGCCTTAAAGC NNNNNNNNNNNNNNN TGGAATTCTCGGGTGCCAAGG AACTCCAGTCAC NNNNNNNNATCTCGTATGCCGTCTTCTGCTTG -3' <--------------ACCTTAAGAGCCCACGGTTCCTTGAGGTCAGTG